Articles
- Page Path
- HOME > J Prev Med Public Health > Volume 45(1); 2012 > Article
-
Special Article
Molecular Typing in Public Health Laboratories: From an Academic Indulgence to an Infection Control Imperative - Franz Allerberger
-
Journal of Preventive Medicine and Public Health 2012;45(1):1-7.
DOI: https://doi.org/10.3961/jpmph.2012.45.1.1
Published online: January 31, 2012
Austrian Agency for Health and Food Safety (AGES), Vienna, Austria.
- Corresponding author: Franz Allerberger, MD. Spargelfeldstr. 191, A-1220 Vienna, Austria. Tel: +43-50555-35500, Fax: +43-50555-95-35500, Franz.Allerberger@ages.at
• Received: October 25, 2011 • Accepted: November 15, 2011
Copyright © 2012 The Korean Society for Preventive Medicine
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
- Using three Austrian case studies, the variegated applications of molecular typing in today's public health laboratories are discussed to help illustrate preventive management strategies relying on DNA subtyping. DNA macrorestriction analysis by pulsed field gel electrophoresis has become the gold standard for subtyping of food borne pathogens like listeria, salmonella, campylobacter and Bacillus cereus. Using a Salmonella Mbandaka outbreak from the year 2010 as example, it is shown how the comparison of patterns from human isolates, food isolates, animal isolates and feed isolates can allow to identify and confirm a source of disease. An epidemiological connection between the simultaneous occurrence of tuberculosis in cattle and deer with cases of human tuberculosis due to Mycobacterium caprae in 2010 was excluded using mycobacterial interspersed repetitive units variable-number tandem repeats subtyping. Also in 2010, multilocus sequence typing with nonselective housekeeping genes, the so-called sequence based typing protocol, was used to elucidate connections between an environmental source (a hospital drinking water system) and a case of legionellosis. During the last decades, molecular typing has evolved to become a routine tool in the daily work of public health laboratories. The challenge is now no longer to simply type microorganisms, but to type them in a way that allows for data exchange between public health laboratories all over the world.
- In 1978 when the Nobel Prize in physiology and medicine was awarded jointly to a German, Werner Arber, and the two Americans, Daniel Nathans and Hamilton O. Smith, "for the discovery of restriction enzymes and their application to problems of molecular genetics," public health laboratories in Europe were essentially still structured in the same way as when they were originally founded. In the late 1890s Robert Koch had initiated routine testing of stool specimens to detect human carriers of Salmonella Typhi [1]. Screening for Corynebacterium diphtheriae, for Streptococcus pyogenes (the causative agent of scarlet fever, then a feared infection) and Mycobacterium tuberculosis were other major tasks for public health laboratories, at a time when eradicating pathogens by detecting and isolating human carriers was considered feasible. In the following decades, subtyping, the characterization of microorganisms beyond the species level was restricted to exceptional situations, with results often coming in months after the actual problem had occurred.
- However, infectious disease challenges are manifold and not of static nature. Identifying human carriers is no longer the main task of public health laboratories. The changes in infectious diseases are the result of ongoing changes in human demographics and behaviours, of changes in technology and industry, changes in economic development and land use, increasing and rapid international travel and commerce, ongoing microbial adaptation, and also the consequence of public health measures [2]. Addressing infectious diseases unabatedly requires a strong public health infrastructure to detect and rapidly respond to these emerging threats to health. Identifying clones of pathogens quickly and reliably is now of crucial importance if public health authorities are to respond effectively to the threat posed by a specific infectious source. One of the most notable benefits resulting from the discovery of restriction enzymes is the ability of the new armoury of molecular methods to characterise and quickly identify bacterial clones. In the last three decades, DNA fingerprinting thchniques have become an indispensable tool in the everyday work of public health laboratories; restriction enzymes were the key to this innovation. For the public health laboratory system in Austria (total population 8.3 million), the implementation of these new methods has effectuated a complete restructuring, with a switch from geographic allocation of tasks (six laboratories each serving a certain geographic area) to allocating special tasks to the only two remaining laboratories, each offering specialized service to all of the country. In this special article, the author discusses the variegated applications of molecular typing in today's public health laboratories by presenting three case studies to help illustrate preventive management strategies relying on subtyping. These examples originate from the daily routine work of the Austrian Agency for Health and Food Safety (AGES). AGES is an independent statutory agency established by the Austrian Health and Food Safety Act in 2002. The Austrian examples presented here are from the years 2010/2011 and are supposed to give an overview of the diverse fields of applications of various DNA subtyping methods routinely employed in everyday work.
INTRODUCTION
- Restriction fragment length polymorphism (RFLP) analysis is a direct result of the work of W. Arber, D. Nathans and H.O. Smith. DNA macrorestriction analysis by pulsed field gel electrophoresis (PFGE), which revolutionized precise separation of DNA fragments greater than 40 kb, has even become the gold standard for subtyping of food borne pathogens like listeria, salmonella, campylobacter and Bacillus cereus [3,4]. D.C. Schwartz and C.R. Cantor developed this variation of agarose gel electrophoresis in which the orientation of the electric field across the gel is changed periodically ("pulsed") rather than kept constant as it was in conventional agarose gel electrophoresis [5]. This technology separates large fragments of unsheared microbial chromosomal DNA obtained by embedding intact bacteria in agarose gel plugs, enzymatically lysing the cell wall and digesting the cellular proteins. The intact DNA is digested with an infrequently cutting restriction enzyme. Subsequent restriction fragment length polymorphism analysis allows differentiation of clonal isolates from unrelated ones [6].
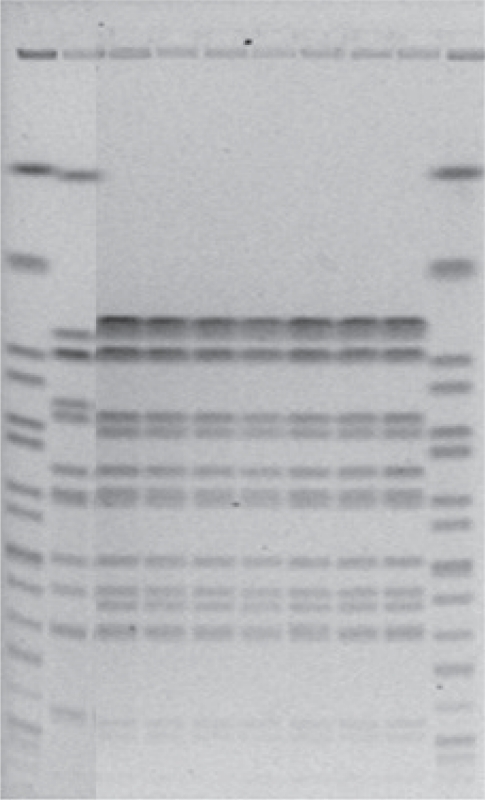
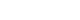
- In April 2010, the Austrian reference centre for salmonella noticed a sudden increase in the number of salmonelloses due to a usually rare serotype: while only five human Salmonella enterica subsp. enterica serovar Mbandaka-infections were documented in the year 2009, 19 such cases were noticed during the two weeks from March 10 to March 31. Isolates were subjected to molecular analysis using PFGE [7,8] and isolates from the year 2010 were found to be indistinguishable from each other but clearly different from those received in previous years. Epidemiological investigation revealed consumption of raw or insufficiently heated eggs or egg dishes as the highly likely source of infection. Active case findings finally resulted in 159 microbiologically confirmed cases. A leftover egg sample from a patient's refrigerator tested positive for Salmonella Mbandaka. Because the epidemiologically incriminated eggs were traced to 56 different Austrian producers but no increase was noticed in neighbouring countries, like Germany or Italy, 226 laying hen flocks serving the 56 producers were environmentally sampled. Although three months had passed between the beginning of the outbreak and sampling of the farms, faecal dust samples from two laying hen flocks still tested positive for the outbreak strain, as did a specimen from a commercial feed product taken at one of these two farms. From the farm with the positive feed sample, 290 consecutive eggs were tested using pools of five eggs each (testing separated for surface and egg content): Salmonella Mbandaka was cultured from one of 58 tested pools on egg surface only. The investigation proved that consumable eggs were the vehicle of this outbreak, and that Salmonella Mbandaka was introduced into the laying hen flocks involved - from 8 of the 9 Austrian provinces - via commercial feed. Further investigations, with microbiological testing of 226 additional laying hen flocks and tracing their feed supply, finally identified one Austrian feed mill as the sole source of the incriminated feed. Figure 1 presents the PFGE patterns of the outbreak clone using the restriction enzyme XbaI in comparison with isolates of sporadic cases gained in previous years; Salmonella Braenderup H9812, which gives rise to a broad spectrum of precisely defined bands, was used as internal standard. The comparison of PFGE patterns from human isolates, food isolates, animal isolates and feed isolates allowed us to identify and confirm the sources of disease. Once an epidemiological investigation has identified a potential source, PFGE can help confirm that the outbreak strain is found in some stage of food production. This leads to a better understanding of the means of contamination and spread and therefore to better control interventions. However, the time required to complete the procedure (>1 day), the need for relatively specialized equipment for electrophoresis and the fact that PFGE generates band patterns that are difficult to share between laboratories are major disadvantages inherent in this method. The analysis of banding patterns can be time-consuming, and the interpretation of banding patterns is somewhat subjective. As a result, improved methods are being sought, but as yet no single method has been able to replace PFGE.
PULSED FIELD GEL ELECTROPHORESIS
- Ten years ago, gel based RFLP analysis using the repetitive insertion sequence IS6110 as a probe was still considered the gold standard for typing of mycobacterial strains [9]. However, the technical steps of IS6110 RFLP were both labor-intensive and lengthy [10]. Moreover, sophisticated computer image analysis software was required for fingerprinting in large-scale analysis. Therefore, typing of mycobacteria has nearly completely switched from RFLP analysis to a polymerase chain reaction-based typing method that relies on the analysis of tandem repeats present in the Mycobacterium tuberculosis complex-genome in up to 41 genetic elements. These genetic elements, called mycobacterial interspersed repetitive units (MIRUs), are scattered throughout the genome, with a variable number of copies of the repeat unit in each locus. MIRU-variable-number tandem repeats (VNTR) analysis generates numerical values that can easily be shared between laboratories [11].The availability of whole-genome sequences has facilitated the discovery of these VNTRs, loci that contain short strings of nucleotides that are repeated a few to many times. This has led to the birth of this new subtyping method, also called multilocus VNTR analysis (MLVA) [12]. Results are depicted by numerical codes, giving the number of MIRU alleles at each locus tested. The following example shows the potential of this molecular subtyping method to disprove an epidemiological connection. The simultaneous occurrence of tuberculosis in cattle and deer in western Austria and cases of human tuberculosis in eastern Austria were noted in early 2011 and prompted an investigation to elucidate a possible connection.
- Mycobacterium caprae, a recently defined member of the Mycobacterium tuberculosis complex, causes tuberculosis among animals and, to a limited extent, in humans in several European countries [13]. While in the ten year period until 2009, there was only one human tuberculosis case with Mycobacterium caprae documented in Austria (in 2008), in 2010 three such cases were registered for this mandatorily reportable disease. Already in fall 2008, the western provinces Tyrol and Vorarlberg had begun to test all cattle for tuberculosis, after documenting occurrence of Mycobacterium caprae in some herds of one of nine Tyrolean districts [14,15]. More than 100 cows from 75 farms had to be culled. Cattle were supposed to have become infected after grazing on alpine pastures contaminated by diseased deer. In the affected district, the county of Reutte, neighbouring the province of Vorarlberg, more than 10% of the red deer (Cervus elaphus) population was found to be infected with Mycobacterium caprae. Overstocking and winter feeding (with "unnatural" crowding of deer at few feeding places) was considered the cause. In 2010, public authorities ordered decimation of half of approx. 1100 red deer in the affected county. The sudden emergence of three human Mycobacterium caprae infections in 2010 raised the question as to whether there was an epidemiological link between animal and human illness. Figure 2 depicts the geographic origin of the Mycobacterium caprae infections in cattle, deer (unpublished AGES data) and humans documented in 2010 [16-18]. MIRU genotyping was performed on these animal and human Mycobacterium caprae isolates. MIRU patterns from deer and cattle isolates were indistinguishable from each other [15], but clearly different from the three patterns of the human isolates. The molecular genetic analysis did not reflect patterns indicative of spread of Mycobacterium caprae from the veterinary outbreak in western Austria to the human cases in eastern Austria. MIRU-VNTR results were substantiated by epidemiological findings: patient A, a 13 year old girl of Turkish ancestry living in Vienna probably became infected when visiting her grandparents in rural Turkey during summer breaks. Patient B, a 85-year-old lady from Lower Austria, probably got infected by her late husband, who had the cattle of his farm culled due to bovine tuberculosis in the early 1960s. Patient C, a 70-year-old patient from Upper Austria probably got infected around 1965, when the cattle of his farm had to be culled by public order because of tuberculosis. Molecular typing was not only able to exclude a connection between the recent re-emergence of tuberculosis in cattle and deer with the occurrence of human Mycobacterium caprae cases but even to disprove the existence of a human outbreak.
- Ten years ago, a single-endonuclease, amplified fragment length polymorphism analysis method by which the patterns are resolved by standard agarose electrophoresis, was adopted as an international standard by the European Working Group for Legionella Infections (EWGLI), especially for cases of travel-associated legionellosis [19]. However, while this method allowed relatively reliable screening of isolates within a single laboratory, inter-laboratory comparison of the results still posed a significant hurdle. Therefore, EWGLI developed a sequence-based typing (SBT) scheme for clinical and environmental isolates of Legionella pneumophila, which is presently widely used in Europe in the investigation of outbreaks of legionellosis caused by Legionella pneumophila. The advantages of using a multilocus sequence typing approach with nonselective housekeeping genes has been well documented for various microorganisms [20]. Using the so-called SBT protocol, the SBT database (provided by EWGLI in conjunction with the London based Health Protection Agency and the European Centre for Disease Prevention and Control in Stockholm under http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php) allows assignment of the seven ordered alleles, flaA, pilE, asd, mip, mompS, proA, and neuA as described by Gaia et al. [21] and Ratzow et al. [22], and representation as one of presently 1032 sequence types (ST). ST 81 for instance has the allelic profile, i.e. the ordered string of allele numbers separated by commas, 2,10,3,28,9,4,9.
- The following example on Legionella pneumophila underlines the considerable potential of this molecular typing method to elucidate connections between an environmental source, in this case a hospital drinking water system, and a case of human illness. At the end of July 2010, the Austrian reference centre for legionella learned about a case of legionella pneumonia in a 70 year old female patient, who had died on July 26. Onset of symptoms had occurred on July 22, admission to hospital C on July 24. A urinary antigen test performed on day 2 of hospitalization was positive for Legionella pneumophila. The patient, who also suffered from neoplasm bronchi with brain metastases, had previously been hospitalized in hospital A for chemotherapy from July 6 to 16, and after spending four days at her private home (from July 16 to 20) - in hospital B for γ-knife intervention (July 20 to 21). In order to identify the environmental source of infection, efforts were made to obtain a patient isolate for comparison with possible future environmental isolates. A blood culture drawn on July 25 (with a negative result reported by the laboratory of hospital C) was the sole specimen available (autopsy was denied due to religious constraints). Two blood culture bottles were sent to the national reference laboratory and volumes of 0.1 and 0.5 mL were directly plated onto buffered charcoal yeast extract agar with and without glycine, vancomycin, polymycin B and cycloheximide supplementation (Oxoid, Cambridge, UK). After incubating for five days, approximately ten colony forming units (CFU) per ml blood culture medium were gained: Legionella pneumophila serogroup (sg) 1 (ST 81) and Legionella pneumophila sg 3 (ST 93). Double infection is a rare, but not unusual phenomenon, be it in salmonellosis, in tuberculosis or in legionellosis [23]. For the health administrators in charge, an available isolate usually initiates an attempt to identify the source of the patient's infection in order to prevent further fatalities. Legionella is often found in warm and cold water systems of buildings; without proof of clonal identity, the mere demonstration of Legionella pneumophila in a water system cannot be regarded as proof of causal relation with illness. Within the incubation period of 2 to 10 days, the patient had stayed in two hospitals and in her own apartment. The testing of water samples, drawn from the patient's room in hospitals A and B and at her home, demonstrated legionella contamination in all three facilities. Water samples (cold and hot water, mixed) obtained on July 29 from the room occupied by the patient during her stay in hospital A yielded 1 CFU Legionella pneumophila sg 1 (ST 81)/100 mL from the sample "hand washbasin tap" and 7 CFU Legionella pneumophila sg 3 (ST 93)/100 mL from the sample "shower". Water samples (again cold and hot water, mixed) obtained on July 29 from the room occupied by the patient during her stay in hospital B yielded 1 CFU Legionella pneumophila sg 1 (ST 442)/100 mL from the sample "shower". Water samples obtained on August 4 from the patient's home yielded 1-220 CFU Legionella pneumophila sg 10/100 mL. Sequence based typing revealed that hospital A was the causative reservoir. The financial resources necessary to sanitize this contaminated drinking water system would not have been allocated without microbiological proof of a causal connection with human illness.
SEQUENCE BASED TYPING
- Molecular typing of pathogens causing diseases complements the traditional epidemiological surveillance by providing appropriate discriminatory analyses to foster rapid and early detection of dispersed clusters or outbreaks, and for detection and investigation of transmission chains. It also supports studies to trace-back the source of an outbreak and to identify new risk factors as the strains can be linked more accurately to epidemiological and clinical data. All of this information can be applied towards improving and better targeting existing infectious disease prevention and control measures and thus presents a clear and immense benefit for the public health and public health policies.
- Molecular typing has evolved to become a routine tool in the daily work of public health laboratories. As this holds true not only for Austria, but also for many other European member states, extensive work is presently being done to prepare for a molecular infectious diseases surveillance system in the European Union [24]. Starting with food and waterborne diseases and with tuberculosis, the ECDC in Stockholm, Sweden, focuses on establishing centralised databases for typing results from PFGE and from MLVA. In the case of food and waterborne diseases, this will enable the linkage of national sporadic cases or outbreaks across the European member states' borders and beyond. The system will start with nontyphoidal Salmonella, enterohaemorrhagic Escherichia coli and Listeria monocytogenes and it will be compatible with the global surveillance of food and waterborne diseases-system led by the Centers for Disease Control and Prevention in Atlanta, Georgia (PulseNet International) and the World Health Organization in Geneva, Switzerland (Global food-borne infections network). To enhance capacity building in the European member states, an MLVA implementation project has already been initiated with the aim to support establishment of the methodology in Mycobacterium tuberculosis reference laboratories. To enhance the capacity for PFGE methodology, a first hands-on workshop took place in June 2011 in order to support Listeria monocytogenes molecular typing implementation as a prerequisite for effective surveillance systems. So the challenge is now no longer to simply type microorganisms, but to type them in a unified way that allows for data exchange between public health laboratories all over the world. To accomplish that, a significant investment in bioinformatics will be required in order to translate large amounts of metagenomics (gene-based) data into a form that provides useful epidemiologic information. New DNA sequencers can determine more than 100 megabases of DNA sequences per run, and new sequencing technologies eliminate the bacterial cloning step used in traditional Sanger sequencing. Instead, they multiply single isolated DNA molecules and analyze them with computers capable of massive parallel processing [25]. The emphasis of future DNA subtyping developments should be on the use of sequence based metagenomics rather than on the gel based methods still used in many public health laboratories. The end-point of typing techniques will be sequencing a whole genome of a pathogen which has the highest discriminatory power [26,27]. However, - as illustrated by the examples of molecular typing presented in this manuscript - for epidemiologic purposes for many diseases methods with lower discriminatory power are presently the gold-standard for public health purposes.
- Addressing emerging infectious diseases requires international and interdisciplinary partnerships to assemble an appropriate infrastructure to detect and respond to these threats to health. Sequence-based typing methods are offering new perspectives of enhanced resolution and comparability of typing systems for public health applications. As gene sequencing technology develops further and methods of sequence analysis become even more user-friendly, new typing methods will evolve and promote acceptance of routine molecular typing in public health laboratories even more.
CONCLUSION
ACKNOWLEDGEMENTS
-
The author has no conflicts of interest with the material presented in this paper.
-
This article is available at http://jpmph.org/.
Notes
- 1. Berger S. Bakterien in Krieg und Frieden: eine Geschichte der medizinischen Bakteriologie in Deutschland 1890-1933. 2009. Gottingen: Wallstein Verlag; p. 143-170 (German)
- 2. Cohen ML. Resurgent and emergent disease in a changing world. Br Med Bull 1998;54(3):523-532. 10326281ArticlePubMed
- 3. Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytia-Trees E, et al. PulseNet USA: a five-year update. Foodborne Pathog Dis 2006;3(1):9-19. 16602975ArticlePubMed
- 4. Choi KB, Lim HS, Lee K, Ha GY, Jung KH, Sohn CK. Epidemiological investigation for outbreak of food poisoning caused by Bacillus cereus among the workers at a local company in 2010. J Prev Med Public Health 2011;44(2):65-73. (Korean). 21483225ArticlePubMedPDF
- 5. Schwartz DC, Cantor CR. Separation of yeast chromosomesized DNAs by pulsed-field gradient gel electrophoresis. Cell 1984;37(1):67-75. 6373014ArticlePubMed
- 6. Graves LM, Swaminathan B, Hunter SB. In: Ryser ET, Marth EH, editors. Subtying Listeria monocytogenes. Listeria, listeriosis and food safety. 2007. 3rd ed. New York: CRC Press; p. 283-304Article
- 7. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006;3(1):59-67. 16602980ArticlePubMed
- 8. Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol 2005;43(3):1045-1050. 15750058ArticlePubMedPMC
- 9. Vincent V, Brown-Elliott BA, Jost KC, Wallace RJ. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. Mycobacterium: phenotypic and genotypic identification. Manual of clinical microbiology. 2003. 8th ed. Washington DC: ASM Press; p. 560-584
- 10. Van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 1993;31(2):406-409. 8381814ArticlePubMedPMCPDF
- 11. Sougakoff W. Molecular epidemiology of multidrug-resistant strains of Mycobacterium tuberculosis. Clin Microbiol Infect 2011;17(6):800-805. 21682800ArticlePubMed
- 12. Lindstedt BA. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 2005;26(13):2567-2582. 15937984ArticlePubMed
- 13. Prodinger WM, Brandstatter A, Naumann L, Pacciarini M, Kubica T, Boschiroli ML, et al. Characterization of Mycobacterium caprae isolates from Europe by mycobacterial interspersed repetitive unit genotyping. J Clin Microbiol 2005;43(10):4984-4992. 16207952ArticlePubMedPMC
- 14. Nindler P, Paschinger A. Bauern fordern Tbc-Freiheit: Rotwild vor Totalabschuss. Tiroler Tageszeitung. 2011. cited 2011 Dec 20. Available from: http://www.tt.com/csp/cms/sites/tt/Tirol/2448885-2/bauern-fordern-tbc-freiheit--rotwild-vor-totalabschuss.csp (German)
- 15. Rotwild im AuBerfern von TBC befallen: MaBnahmen notwendig. Tiroler Tageszeitung. 2011. cited 2011 Dec 20. Available from: http://www.tt.com/csp/cms/sites/tt/%C3%9Cberblick/Chronik/ChronikTirol/2452482-6/rotwild-im-au%C3%9Ferfern-von-tbc-befallen--ma%C3%9Fnahmen-notwendig.csp (German)
- 16. Salgado-Voss AS, Much P, Rendi-Wagner P, Herzog U. Bericht uber Zoonosen und ihre Erreger in osterreich im Jahr. 2010. cited 2011 Dec 20. Available from: http://www.ages.at/uploads/media/Zoonosenbroschuere_2010.pdf (German)
- 17. Amt der Vorarlberger Landesregierung. Resume zum Jagdjahr. 2010/2011. cited 2011 Dec 20. Available from: http://www.vorarlberg.at/vorarlberg/landwirtschaft_forst/landwirtschaft/jagd/neuigkeiten_mitbild_/r_sum_zumjagdjahr2010_201.htm (German)
- 18. Amt der Tiroler Landesregierung. Tbc im Oberen Lechtal gemeinsam bekampfen. cited 2011 Dec 20. Available from: http://www.tirol.gv.at/presse/meldungen/meldung/artikel/tbc-im-oberen-lechtal-gemeinsam-bekaempfen/?no_cache=1&cHash=a75c9d6810 (German)
- 19. Fry NK, Bangsborg JM, Bergmans A, Bernander S, Etienne J, Franzin L, et al. Designation of the European Working Group on Legionella Infection (EWGLI) amplified fragment length polymorphism types of Legionella pneumophila serogroup 1 and results of intercentre proficiency testing using a standard protocol. Eur J Clin Microbiol Infect Dis 2002;21(10):722-728. 12415471ArticlePubMed
- 20. Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol 1999;7(12):482-487. 10603483ArticlePubMed
- 21. Gaia V, Fry NK, Afshar B, Luck PC, Meugnier H, Etienne J, et al. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol 2005;43(5):2047-2052. 15872220ArticlePubMedPMC
- 22. Ratzow S, Gaia V, Helbig JH, Fry NK, Luck PC. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J Clin Microbiol 2007;45(6):1965-1968. 17409215ArticlePubMedPMC
- 23. Pavlic M, Allerberger F, Dierich MP, Prodinger WM. Simultaneous infection with two drug-susceptible Mycobacterium tuberculosis strains in an immunocompetent host. J Clin Microbiol 1999;37(12):4156-4157. 10565951ArticlePubMedPMCPDF
- 24. European Centre for Disease Prevention and Control. Annual report of the director 2010. 2011. Stockholm: European Centre for Disease Prevention and Control; p. 13
- 25. Hoorfar J, Christensen B, Pagotto F, Rudi K, Bhunia A, Giffiths M. In: Hoorfar J, editor. Future trends in rapid methods: where is the field moving, and what should we focus on? Rapid detection, characterization, and enumeration of foodborne pathogens. 2011. Washington DC: ASM press; p. 413-420Article
- 26. Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science 2011;331(6016):430-434. 21273480ArticlePubMedPMC
- 27. He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A 2010;107(16):7527-7532. 20368420ArticlePubMedPMC
REFERENCES
Figure 1
Pulsed-field gel electrophoresis patterns using the restriction enzyme XbaI.
Lanes 1 and 10: Salmonella Braenderup (internal standard), lane 2: human Salmonella Mbandaka isolate from 2010, epidemiologically not related to the 2011-outbreak, lanes 3-9: Salmonella Mbandaka outbreak clone (lanes 3-6: isolates from outbreak cases, lane 7: isolate from an epidemiologically involved left over egg, lane 8: isolate from a faecal dust sample of an epidemiologically involved laying hen flock, lane 9: isolate from the causative feed).
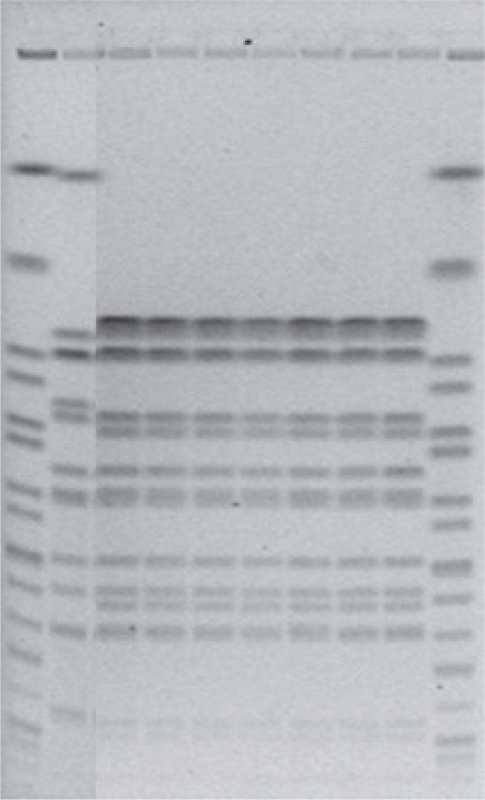

Figure & Data
References
Citations
Citations to this article as recorded by 

- Ground water as the source of an outbreak of Salmonella Enteritidis
Ana Kovačić, Željko Huljev, Edita Sušić
Journal of Epidemiology and Global Health.2017; 7(3): 181. CrossRef - Distribution of Salmonella serovars along the food chain in Poland, 2010–2015
Magdalena Skarżyńska, Andrzej Hoszowski, Magdalena Zając, Anna Lalak, Ilona Samcik, Renata Kwit, Dariusz Wasyl
Journal of Veterinary Research.2017; 61(2): 173. CrossRef - The risk of carriage of Salmonella spp. and Listeria monocytogenes in food animals in dynamic populations
Korana Stipetic, Yu‐Chen Chang, Kenlyn Peters, Ahmed Salem, Sanjay H. Doiphode, Patrick L. McDonough, Yung Fu Chang, Ali Sultan, Hussni O. Mohammed
Veterinary Medicine and Science.2016; 2(4): 246. CrossRef - Molecular typing of bacteria for epidemiological surveillance and outbreak investigation / Molekulare Typisierung von Bakterien für die epidemiologische Überwachung und Ausbruchsabklärung
Werner Ruppitsch
Die Bodenkultur: Journal of Land Management, Food and Environment.2016; 67(4): 199. CrossRef - Legionella detection and subgrouping in water air-conditioning cooling tower systems in Kuwait
Qadreyah Al-Matawah, Sameer Al-Zenki, Ahmad Al-Azmi, Tahani Al-Waalan, Fadila Al-Salameen, Ahmad Ben Hejji
Environmental Science and Pollution Research.2015; 22(13): 10235. CrossRef - Listeriosis cluster in Sydney linked to hospital food
Zeina Najjar, Leena Gupta, Vitali Sintchenko, Craig Shadbolt, Qinning Wang, Narinder Bansal
Medical Journal of Australia.2015; 202(8): 448. CrossRef - Diversity of pulsed-field gel electrophoresis patterns of cereulide-producing isolates ofBacillus cereusandBacillus weihenstephanensis
Virginie Castiaux, Elise N'Guessan, Izabela Swiecicka, Laurence Delbrassinne, Katelijne Dierick, Jacques Mahillon
FEMS Microbiology Letters.2014; 353(2): 124. CrossRef - Mycobacterium capraeinfection in humans
Wolfgang M Prodinger, Alexandra Indra, Orhan K Koksalan, Zeki Kilicaslan, Elvira Richter
Expert Review of Anti-infective Therapy.2014; 12(12): 1501. CrossRef - Same-Day Subtyping of Campylobacter jejuni and C. coli Isolates by Use of Multiplex Ligation-Dependent Probe Amplification–Binary Typing
Angela J. Cornelius, Olivier Vandenberg, Beth Robson, Brent J. Gilpin, Stephanie M. Brandt, Paula Scholes, Delphine Martiny, Philip E. Carter, Paul van Vught, Jan Schouten, Stephen L. W. On, D. J. Diekema
Journal of Clinical Microbiology.2014; 52(9): 3345. CrossRef - Strukturelle Voraussetzungen und Bedingungen für eine effektive mikrobiologische Diagnostik bei Ausbruchsgeschehen
F. Allerberger
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz.2013; 56(1): 22. CrossRef
 KSPM
KSPM


 PubReader
PubReader ePub Link
ePub Link Cite
Cite